1.The papers I present are NOT my own reasoning, fucking deal with it.spinoza99 wrote:It is very hard to have a real dialogue with you. It is obvious from the profanity that you harbor such a burning hatred for my ideas that even if you did find a flaw in you're own reasoning you would never admit to it. It would be irrational for me to expect to learn anything from someone who would never admit to a truth that ran counter to his beliefs.
2. It would be rational for you to learn from the literature and ensuring you can actually understand elementary biology before you go spouting crap off.
3. The scientific consensus is NOT a belief, projection much?
4. If it is so hard for you to have a dialogue with me I suggest you learn to read by yourself, or go to Uni and get an education first.
5. I will change my stance as and when papers pass through scientific peer review in proper journals, if you have such evidence, present it, publish or perish, put up or shut the fuck up.
6. I am not expecting you to learn anything from me, the free explanations of a body of well established scientific literature are for the passers by who may be swayed by such guff as has been forthcoming from you thanks to ignorance, I do not expect you to learn anything at all since it is clear that you do not have the scientific knowledge required to even begin to appreciate the papers I'm presenting here, it isn't all about you, deal with it.
7. Belief is such a fuckwitted word, the scientific community doesn't operate on faith you know, if evidence does pop up in peer reviewed literature I'm willing to change my stance, insisting upon high standards of evidence =/= being close minded, your handwaving of reality because it gives your mythology a middle finger IS, and any attempts to suggest otherwise is nothing more than a blatant case of projection.
8. "You are biased, and rude, and hard to speak to, therefore intelligence causes mutations and therefore randomness cannot be involved" What kind of non-sequitur is that?
They chose 139 random polypeptides,
They being the scientists, there are bioinformatics tools that generate random peptide sequences, such as RandSeq which generates a random protein sequence, which can be found on Expasy's server.Who is they? We're talking about 20 different amino acids and they all have to line up in a precise order. How does one amino acid know to get in the right order?
1) Generate sequence.
2) Convert sequence to DNA (amino acids are mapped to specific nucleotide triplets)
3) Synthesize DNA
4) Insert DNA into phage genomes by genetic engineering using a plasmid.
In this case they introduced an external random sequence to ensure they'd start off with a random sequence and that there was no co-optation involved.
How do the amino acids know how to line up?
Translation. (Again elementary biology)
The chemistry of which is represented thus
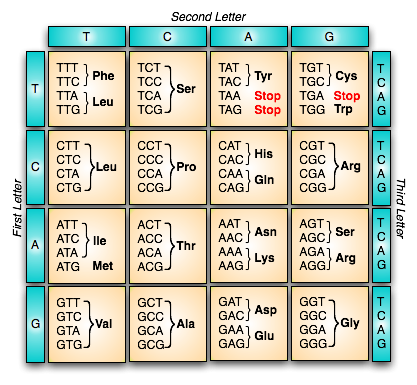
And the process of which occurs thus

they synthesized the DNA corresponding to that, they introduced that into bacteriophages, rendering them defective.
Substitutions by random mutation during phage replication began to act on that sequence, changes in sequence resulted in improving fitness (an increase in infectivity)
Fucking nonsense, there are proteins whose functions (as in efficiency of functioning) vary, for the better and for the worse. A random sequence of 300 nucleotides will produce a random protein of 100 amino acids.Proteins are not fit, they either work or they don't, it's binary, 1 or 0. You can't have a protein with less than a 100 or amino acids.
proteins have shapes, conferring function, different shapes have different functions and this functionality can vary, and what citation do you bring to support your assertion? Precisely none. I will also present another paper to show you how asinine your assertion is.
The evidence is here and by the look of things the full paper is downloadable.
http://www.springerlink.com/content/32vn61n3h463k270/The large subunit (HycE, 569 amino acids) of Escherichia coli hydrogenase 3 produces hydrogen from formate via its Ni–Fe-binding site. In this paper, we engineered HycE for enhanced hydrogen production by an error-prone polymerase chain reaction (epPCR) using a host that lacked hydrogenase activity via the hyaB hybC hycE mutations. Seven enhanced HycE variants were obtained with a novel chemochromic membrane screen that directly detected hydrogen from individual colonies. The best epPCR variant contained eight mutations (S2T, Y50F, I171T, A291V, T366S, V433L, M444I, and L523Q) and had 17-fold higher hydrogen-producing activity than wild-type HycE. In addition, this variant had eightfold higher hydrogen yield from formate compared to wild-type HycE. Deoxyribonucleic acid shuffling using the three most-active HycE variants created a variant that has 23-fold higher hydrogen production and ninefold higher yield on formate due to a 74-amino acid carboxy-terminal truncation. Saturation mutagenesis at T366 of HycE also led to increased hydrogen production via a truncation at this position; hence, 204 amino acids at the carboxy terminus may be deleted to increase hydrogen production by 30-fold. This is the first random protein engineering of a hydrogenase.
Sequence variations introduce different levels of functioning, and when placed under selection, for example in microbes which use H2S for metabolism, as an electron acceptor, those that produce more hydrogen will have more fitness than those that produce less, protein function IS not digital, wherefrom you got such a mistaken understanding I do not know, unless you are deliberately spewing forth falsehoods to provoke, in which case you are nothing more than a despicable troll.
Of course, see the second graph on the PLoS paper, where the functional efficiency, in this case infectivity, increased with substitution and selection also lays waste to your assertions, there are not just two states of function, but various states that climb steadily, in other words, it is NOT binary. Evidence says you are wrong, so deal with it, and if you don't it is you and not me who will ignore anything that goes against your view regardless of the standard of evidence, which unsurprisingly like most people who project their character onto others, you are.
This is so wrong I ddon't know where to start.With a sequence 120 long made up 20 different amino acids, the sum total of possible sequences is 10^150 roughly. There are about 600 different proteins. Just to get the odds down to 10^80 of one protein forming you would need 10^70 different possible protein sequences. There of course may be more 600 different proteins, but anything more than 2000 is highly unlikely, 10,000 impossible.It will also show you that to have a functioning phage protein there is more than one possible sequence, firstly , and secondly, mutation + selection shape random sequences into functional ones , your argument of "139 precise order 139 precise order" is nothing more than a fuckwitted exposition of dimwitted ignorance of elementary molecular biology.
1.Strawman...proteins just don't pop into existence out of nowhere, read about translation.
2.There are 20,000 proteins in humans.
3.As for asking where they can come from, try gene duplication and whole genome duplication. Proteins are produced by the translation of genomes, genomes are produced and acted upon by mutagenic processes. Mutagenic processes can generate proteins with diverse functions from genomic material. Natural processes explain the evolution of proteins, now that is it.
I am half intending to report you for trolling.
PS- "You cannot have a protein that is less than" assertion has this in reply.
There is an example of a very short polypetide that assume 3d comformation, which is the pre-requisite for protein function.
http://www.cell.com/structure/retrieve/ ... 2604002424We have designed a peptide termed chignolin, consisting of only 10 amino acid residues (GYDPETGTWG), on the basis of statistics derived from more than 10,000 protein segments. The peptide folds into a unique structure in water and shows a cooperative thermal transition, both of which may be hallmarks of a protein. Also, the experimentally determined β-hairpin structure was very close to what we had targeted. The performance of the short peptide not only implies that the methodology employed here can contribute toward development of novel techniques for protein design, but it also yields insights into the raison d'etre of an autonomous element involved in a natural protein. This is of interest for the pursuit of folding mechanisms and evolutionary processes of proteins.
Oxytocin, which also has functions, is a peptide hormone consisting of just nine peptides. (proteins are defined as just polypeptides with a weight of more than 10,000 Daltons)